cbpp 데이터셋
lme4::cbpp()
data(cbpp, package="lme4")'도구 > 패키지 적재하기...' 메뉴 기능을 선택하고 lme4 패키지를 찾아서 선택한다.
그리고 '데이터 > 패키지에 있는 데이터 > 첨부된 패키지에서 데이터셋 읽기...' 메뉴 기능을 선택하면 하위 선택 창으로 이동한다. 아래와 같이 lme4 패키지를 선택하고, cbpp 데이터셋을 찾아서 선택한다.

cbpp 데이터셋이 활성화된다. R Commander 상단의 메뉴에서 < 활성 데이터셋 없음> 이 'cbpp'로 바뀐다.
summary(cbpp)
str(cbpp)'통계 > 요약 > 활성 데이터셋' 메뉴 기능을 통해서 cbpp 데이터의 요약 정보를 살펴보자. str() 함수를 이용하여 cbpp 데이터셋의 내부 구조를 살펴보자.

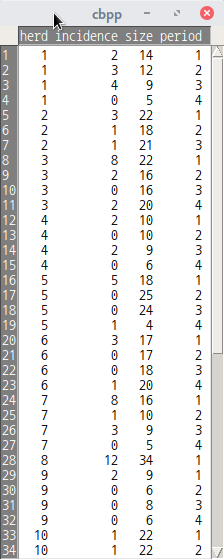
| cbpp {lme4} | R Documentation |
Contagious bovine pleuropneumonia
Description
Contagious bovine pleuropneumonia (CBPP) is a major disease of cattle in Africa, caused by a mycoplasma. This dataset describes the serological incidence of CBPP in zebu cattle during a follow-up survey implemented in 15 commercial herds located in the Boji district of Ethiopia. The goal of the survey was to study the within-herd spread of CBPP in newly infected herds. Blood samples were quarterly collected from all animals of these herds to determine their CBPP status. These data were used to compute the serological incidence of CBPP (new cases occurring during a given time period). Some data are missing (lost to follow-up).
Format
A data frame with 56 observations on the following 4 variables.
herd
A factor identifying the herd (1 to 15).
incidence
The number of new serological cases for a given herd and time period.
size
A numeric vector describing herd size at the beginning of a given time period.
period
A factor with levels 1 to 4.
Details
Serological status was determined using a competitive enzyme-linked immuno-sorbent assay (cELISA).
Source
Lesnoff, M., Laval, G., Bonnet, P., Abdicho, S., Workalemahu, A., Kifle, D., Peyraud, A., Lancelot, R., Thiaucourt, F. (2004) Within-herd spread of contagious bovine pleuropneumonia in Ethiopian highlands. Preventive Veterinary Medicine 64, 27–40.
Examples
## response as a matrix
(m1 <- glmer(cbind(incidence, size - incidence) ~ period + (1 | herd),
family = binomial, data = cbpp))
## response as a vector of probabilities and usage of argument "weights"
m1p <- glmer(incidence / size ~ period + (1 | herd), weights = size,
family = binomial, data = cbpp)
## Confirm that these are equivalent:
stopifnot(all.equal(fixef(m1), fixef(m1p), tolerance = 1e-5),
all.equal(ranef(m1), ranef(m1p), tolerance = 1e-5))
## GLMM with individual-level variability (accounting for overdispersion)
cbpp$obs <- 1:nrow(cbpp)
(m2 <- glmer(cbind(incidence, size - incidence) ~ period + (1 | herd) + (1|obs),
family = binomial, data = cbpp))